A recent reply by the brilliant scientist Darren Naish to one of my posts concluded by saying, “But the fact is you aren’t right: you know full well that the reason we reject your proposals, your hypotheses, your trees, and the observations that all of those things are based on is that you’re using a wholly unreliable technique that cannot be accepted as proper science. You need to stop thinking that everyone is against you because you are heretical. No, it’s because you’re mostly wrong.”
This is pretty harsh criticism, but note that it is lacking in any evidence. Unfortunately, this is typical of what I hear from other paleontologists and frankly, that’s not how scientists should interact. That’s not how I present my hypotheses. I show my work and others should too. And it better be good evidence. Unassailable. If the “unreliable” technique I’m using is using photographs, well, that’s how scientists share their work — and everyone does that.
The fact is, no one has attempted to duplicate my taxon list, even in their own way or even to a minor extent. All I ask is for someone to add a few lizards, especially those I found to nest close to pterosaurs in the large reptile tree (Huehuecuetzpalli, Sharovipteryx, Cosesaurus, etc), to any of the recent studies that nest pterosaurs close to Scleromochlus and phytosaurs. They can use their own observations, their own character traits, etc. Excluding other candidates a priori is not good science, especially when they appear to be the correct candidates and the alternatives all have major problems (as detailed in the last 300 or so blogs) and recognized by many.
I’m not trying to be right. I’m reporting results. If anyone can produce a valid study that produces different results, I’ll be more than happy to report on them. Shying away from such a test is not good for science. I’m happy to get your comments, especially those that indicate where I and others have made mistakes. That’s how we all correct the errors of the past and come to more valid conclusions.
I am constantly finding errors in my work as new data comes in. Yes, that has always been true. But the corrections of those errors have only served to clarify relationships and anatomy.
I haven’t met a phytosaur yet that even vaguely could ever claim to be a pterosaur uncle. Yet that’s the current paradigm that no one wants to talk about. So, let’s put the shoe on the other foot and imagine that everyone knows that pterosaurs are lizards derived from Cosesaurus and kin. Now imagine the furor that would erupt if I were to propose that phytosaurs were the closest cousins of pterosaurs.
The current situation just doesn’t make any sense.
This is the power of the current paradigm. Charles Darwin had to fight his then current paradigm. So did others. It’s time to see pterosaurs and reptiles in a new light, based on rigorous evidence and large taxonomic studies that leave little room for doubt.
Please, someone, anyone, just give it a try.
My best to you all.
___________________________________
Addendum in Response to Darren Naish’s note from Saturday, May 12.
“I’m only going to say one more thing here… so long as you keep saying that people “link phytosaurs with pterosaurs”, it’s obvious that you do not understand how to interpret phylogenetic trees. And I agree with David M about your use of characters and codings.”
How else can we interpret this tree? There is an unknown sister taxon between phytosaurs and erythrosuchids that is the closest sister to pterosaurs, according to this tree. This is the image posted by Darren Naish in his blog Scientific American – Tetrapod Zoology. This tree reflects trees recovered by Nesbitt (2011) and Brusatte et al. (2010) in which no other closer basal sister taxa are provided for pterosaurs.
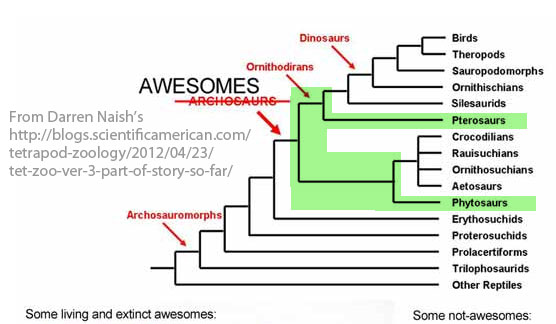
Figure 1. Phytosaurs basal to pterosaurs. Note there are no closer taxa offered at the base of the Pterosauria than the Phytosauria. The Erythrosuchia + the Phytosauria do not include a gradual accumulation of pterosaurian characters. Image attribution listed above.
References
Brusatte SL , Benton MJ , Desojo JB and Langer MC 2010. The higher-level phylogeny of Archosauria (Tetrapoda: Diapsida), Journal of Systematic Palaeontology, 8:1, 3-47.
Nesbitt SJ 2011. The early evolution of archosaurs: relationships and the origin of major clades. Bulletin of the American Museum of Natural History 352: 292 pp.

Dave – thanks for the kind words. In response…. Firstly, the text of mine that you’re referring to was a comment (or two, or three) on a blog, not exactly the place where I’d want to present the evidence you’re referring to. It was something I wanted to write and post in, hopefully, less than three minutes.
Secondly, it would take time and effort to do the job properly and produce a peer-reviewed response to your work. I’m not about to do this, and I note that others aren’t either. Why? Several reasons. One is that everyone is extremely busy publishing their own work, and few are prepared to take the time to knock down a minority opinion (that, let’s face it, hasn’t impacted on the field anyway – I don’t wish to be rude, but your views are universally regarded as ‘fringe’ ones). How scientists should react to fringe, minority opinions is something that gets discussed quite a bit – ideas differ, but many argue that fringe ideas should simply be ignored and allowed to die without any ‘oxygen of citation’.
Thirdly, you keep claiming that people aren’t running the same datasets as you, and that’s why they aren’t getting the same results. No, there are several projects out there where people have sampled widely across reptiles and amniotes (Müller and colleagues, Merck) and they don’t recover phylogenies at all similar to yours. Mickey Mortimer has even made a point of informally testing your claims and he always finds them to be non-supportable (maybe we should push him to publish this work?). And those researchers who publish on this area are looking at actual fossils (not photos) _and_ are able to convince other researchers that what they report actually exists in reality. I’m afraid that I (and others) regard your datasets (and hence your functional and phylogenetic hypotheses) as rotten to the core since they’re based on your photo-tracing technique. You keep justifying this technique as if it’s something that everybody else does, but – no, you are doing something weird, unique, subjective, and utterly unacceptable. You are reporting structures, bones, babies, soft-tissue frills and all manner of other craziness that does not exist, and you continue to argue that those of us who do not see these things as real are refusing to credit that you might sometimes be right. I don’t doubt that you _might_ be right, sometimes (yes, I know full well about the particular _Langobardisaurus_ specimen), the problem is that ALL of your observations incorporate info from your photo-tracing technique. As long as the photo-tracing technique reigns supreme in your work, your body of work as a whole remains unworthy of serious consideration. There also seem to be problems in the way you interpret trees, your ideas about what support statistics mean, and the time and effort involved in building coding data.
To wrap up, my concern these days – and the reason that I’m here on your blog in the first place – is that your stuff has such major ‘web presence’ that you are effectively mis-educating lots of people who search for zoological data online. Naive parties see your reconstructions and hypotheses and think that what you’re reporting is (1) accurate and (2) ‘mainstream’. I’m aiming to do something about this soon, though please be assured that it’s nothing personal. All the best.
Darren, it’s not just the “photointerpretation technique” (pareidolia, as I demonstrated on my unfortunately defunct website – I’m no longer a student, so my webspace is gone).
David once was so kind as to send me an early version of his amniote phylogeny dataset. It has never been published, so I don’t want to talk about it in detail, but it’s safe to say that David had at the time – and likely still has – no clue about how to make a dataset for phylogenetic analysis. The matrix was flooded with correlated characters. Line drawings from the literature were misinterpreted to the extent that pterygoids got misinterpreted as a lower temporal bar that simply isn’t there. And so on and so forth! After a few e-mails back and forth, David withdrew from the conversation.
I plan to start a blog, uh, sometime, I guess. I suppose I can put up my interpreted photos up there.
No, dinosaurs, not phytosaurs.
Really, why do you act as if you hadn’t read the second half of this?
“Just”, or “give it a try”?
As I’ve been telling you for years, that’s a whole PhD thesis you’re talking about. That you’re not treating it as such is a problem for the reliability of your work, not for anything else.
David, Not a whole PhD thesis. Simply add Huehuecuetzpalli, a known lizard, to any current trees that include pterosaurs and see if it nests outside the clade or close to the pterosaurs. Or if the pterosaurs nest outside the clade with the lizard. One taxon is all it will take. Not a big deal.
Regarding earlier matrices, I’ve been correcting errors ever since and continue to do so when I find them. So please get the update if you’re interested.
Regarding phytosaurs, you’ll note that just “south” of pterosaurs you’ll often find proterochampsids and phytosaurs on most trees. Obviously they don’t belong together, which is exactly my point. “North” of pterosaurs you find dinosaurs, as you mentioned, on my published trees.
I understand that you speak for the majority, Darren. Bottom line is, my sisters look like each other. They also produce gradual accumulations of characters found in derived taxa. To have hundreds of taxa and hundreds of characters result in a single, fully resolved tree is the ideal, and I’ve achieved that. Thus my tree appears to echo the natural order. You can’t say that about competing trees that link mesosaurs with pareiasaurs and phytosaurs with pterosaurs, which is the current paradigm. Please send reference to the Merck tree and any others that I have only heard about and not seen.
I’m only going to say one more thing here… so long as you keep saying that people “link phytosaurs with pterosaurs”, it’s obvious that you do not understand how to interpret phylogenetic trees. And I agree with David M about your use of characters and codings.
No, you can’t do that. If you add Huehuecuetzpalli, you must add all positions that H. could have. That means you must expand the published analyses of archosaur phylogeny to an analysis of diapsid phylogeny; you must add several squamates, at least one sphenodontian, and a series of stem-lepido- and -archosauromorphs.
Many published archosaur analyses don’t contain any non-archosauromorphs even as outgroups! If you add a taxon that lies outside the clade represented by the taxon sample, you must automatically get rubbish.
And yes, this means that every archosaur analysis that contains pterosaurs but no lepidosauromorphs has failed to test whether pterosaurs are archosaurs. Only a full analysis of diapsid phylogeny can do that. You’re fully correct about this point – like a stopped clock that is right twice a day.
I’m currently working on finding and fixing all the problems with a large published data matrix. Before my current postdoc, it was a chapter of my PhD thesis; and before that, it was a chapter in the PhD thesis of Damien Germain. Trust me, I know how much work properly done phylogenetics is – even though just adding a taxon to a matrix is a matter of hours, a day at most.
Well, I hope you’re better now at recognizing mistakes than 7 years ago.
There is no south and no north on a cladogram. Cladograms have one axis, not two; that axis is time, which proceeds from the root to the branch tips. That’s it. A cladogram is a mobile; rotating a branch around the axis – in this case the root of that branch – doesn’t change anything. A cladogram consists of nothing but its branching pattern, its topology. Follow the link I provided yesterday and read it already, I don’t want to say everything twice within just weeks.
That is the case in every cladogram, because that’s how the computer programs work.
Whether the similarities between sister-groups are easily visible in the general shape of the animals doesn’t depend on that. Is that what you mean?
You’ve kept saying that for years, and we’ve kept trying to explain why it’s not that simple, and you keep not even reacting to that. You just reassert the same thing again and again.
Often, when an analysis results in a single MPT, that’s a sign the data are either insufficient or even cherry-picked. Often, adding more data decreases resolution.
Having all taxa, all data, and the true model of evolution would result in a single tree. But these conditions are never met. Therefore, the number of MPTs is not a support measure.
The argument from personal incredulity is a logical fallacy.
Yes, meso- and pareiasaurs had a common ancestor that looked much like Milleretta sometime around the beginning of the Permian or even earlier, maybe 30 million years before the first mesosaur and 50 before the first pareiasaur. A lot can happen in that amount of time. Early Eocene bats and whales had a common ancestor just about 10 million years earlier.
Yes, if pterosaurs are archosaurs, ptero- and phytosaurs had a common ancestor that looked much like Turfanosuchus sometime in the first half of the Early Triassic (before Xilousuchus!), perhaps earlier still, some 30 million years before the oldest known ptero- and phytosaurs. A lot can happen in that amount of time.
Unpublished PhD thesis that was only presented once in an SVP talk in 2001. *sigh*
Best reference for Merck so far: http://dml.cmnh.org/2000Jun/msg00136.html (it’s not just 2001, but also others like 1997)
I talked to him in person in 2008. He apologized for still not having published…
To David M’s comment: If you can’t add Huehuecuetzpalli, you can’t add pterosaurs, a clad that is just a guilty of being an outgroup. Everyone in the archosaur camp admits that pterosaurs appear “out of nowhere” with few links to archosaurs. This is why. They’re not archosaurs. Convergence gave them an antorbital fenestra, also seen in Cosesaurus and kin.
And adding quotes around “north” and “south” is a way to say “we’re talking informally now, tongue in cheek” as most clades rise on the page from bottom to top, just like most maps put north at the top. Not that it’s right. It’s just a convention.
Yes, I actually said so.
Still doesn’t automatically make your results right.
Oh, there’s Scleromochlus, and then the two mystery beasts that Atanasov still hasn’t published… does anyone know what’s holding him up?
It’s not like just this one feature is holding them there, you know.
I know – but (1) if you hadn’t mentioned what was in which direction, I wouldn’t even have understood which direction you meant by “north”; (2) you implied there was a meaningful axis that simply isn’t there (see below).
It’s more common, actually, for the time axis to be left-to-right than bottom-to-top.
Now to your addendum:
What utter nonsense! The pterosaurs have a sister-group in that tree: it is the (Silesaurids (Ornithischians (Sauropodomorphs (Theropods, Birds)))) clade. Yes, the whole fucking clade is the sister-group of the pterosaurs!!! How the fuck is it possible that you’ve been reading papers about phylogenetics for about 10 years now and still haven’t understood that sister-groups are any two clades that are joined at one node!?!?!
Yes, I am getting upset. On the one hand, there’s stuff I’ve been explaining to you for seven years now, and you just ignore it; on the other hand, there’s stuff that is so basic that I didn’t even notice you don’t understand it – and neither did you!
The sister-group of the phytosaurs is the (Crocodilians, Rauisuchians, Ornithosuchians, Aetosaurs) clade.
The sister-group of the crocodilians is either “Rauisuchians” or “Ornithosuchians” or “Aetosaurs” or some clade composed of 2 or 3 of those – there’s no way to tell from this polytomy.
The sister-group of “Erythrosuchids” is “Awesomes”.
The sister-group of “Other Reptiles” is “Archosauromorphs”.
Do you get it now? If yes, please tell me the sister-groups of “Ornithischians” and “Proterosuchids” in this tree.
David, if you’re going to hang your hat on semantics, you’re still not going to find the ancestors of pterosaurs within that clade of AWESOMES or diminish the validity of the large phylogenetic analysis. If the sister of Phytosaurs + Crocs thru Aetosaurs is Pterosaurs + Dinosauromorphs, then the opposite must also be true. I only simplified things, as if Crocs thru Aetosaurs and Dinosauromorphs had never evolved or existed. We’re looking for the predecessors of pterosaurs, taxa that share many traits with pterosaurs. Unfortunately, as often pointed out, no archosaur (or AWESOME) comes close.
To your other points Scleromochlus was compared to pterosaurs here on July 14 of 2011. Scleromochlus nests with other bipedal crocs, discussed here on Oct. 23, 2011. Testing has shown that turtles are closer to pterosaurs than is Scleromochlus, covered on July 25, 2011. Atanassov’s (2001, 2002) Pteromimus is a basal langobardisaur, covered on Dec. 23 of 2011, much closer to pterosaurs, indeed. But several taxa, the fenestrsaurs are closer still, covered July 16, 2011. Cosesaurus was the first in this lineage to have a pteroid, a prepubis, a strap-like scapula and stem-like, quadrant-shaped, locked down coracoids, traits shared with pterosaurs. You can see these traits at http://www.reptileevolution.com/cosesaurus.htm. Thanks for writing.
You mean that the sister-group of pterosaurs + dinosauromorphs is phytosaurs + crocs-thru-aetosaurs? Yes, by definition.
That’s not a simplification, it’s plain wrong, and nobody is going to understand it. I didn’t.
When I started writing scientific papers, the first thing my supervisor said to me was “you will be misunderstood – by someone, somewhere, sometime –, so it’s your responsibility to minimize the opportunities for that”.
Well, no and no. We’re trying to answer the question “which currently known animals are the closest known relatives of the pterosaurs”. Obviously, they must have something in common with pterosaurs; they must have more synapomorphies with pterosaurs than with anything else; but that doesn’t mean they must have “many traits” in common at all.
Thanks, I’ll read those posts.
Cosesaurus: photointerpreted. It hurts to look at the photos, because they’re so blurry – they’re descreened. You know what I think of that.
The line between “sc” and “st” is a step in the matrix. “st” is probably not real at all, but if it is, it’s probably the right coracoid. “co” is probably a cervical rib, except that the weird flange you put at one end is a completely arbitrary line drawn through the matrix next to the bone. It’s pareidolia.
On to the others…
Why doesn’t this blog have an ordinary archive? Searching for Pteromimus is easy enough, but the others I was only able to find by searching for Scleromochlus and scrolling a lot.
Pteromimus: can’t say much about it, I still haven’t read the thesis, and you only present the skull reconstruction.
No, you only mention your tree there. But fortunately you include a link to your post from Sept. 14, 2011 which discusses the anatomy of Scleromochlus in some detail. It’s definitely interesting, but you’re not saying Benton did anything wrong except for misorienting the quadrate; am I understanding you right? Because if so, I have trouble imagining that this one feature would cause Scleromochlus to come out where it does in Brusatte et al. (2010) and Nesbitt (2011), analyses that contain plenty of crurotarsans.
BTW, why do you think the articular of S. must have been restricted to the caudal end of the lower jaw? It commonly isn’t; such long retroarticular processes are by no means unheard of.
Your post from July 14, 2011, is such a glorious confusion of “ancestor” and “sister taxon” – even now that I understand what you incorrectly mean by “sister taxon” – that there’s simply no point in talking about it. Sisters can evolve in different directions; they’re not mother and daughter, they’re sisters.
Here you say of the tree at the far right that “no outgroups were employed”. That’s nonsense. If you don’t specify an outgroup, PAUP* treats the first taxon in the list as the outgroup and uses it to root the trees. Have you ever read the manual*? I can send you the pdf. – All four trees look like there are not enough characters to sort that many taxa correctly, and you know what I thought of the last matrix of yours that I saw.
* It’s for 3.1, the last non-beta version that was released, but most of it is still accurate. What’s no longer accurate is mentioned in the command reference for 4.0beta10, which I can also send.
I completely agree with David regarding your phylogenetic terminology and your character list quality. Will be interesting to see what Darren comes up with…
Mickey, I hope you and others will take me up my challenge on May 12 to find the two or more taxa that should not be sisters in my reptile and pterosaur trees and shift the oddball(s) where you think they would be more parsimoniously nested. You don’t have to back up your reasoning, but it would be nice to hear it. Certainly if there’s something wrong here, than the results should not be as they are. Find the bad nestings and let’s work this out.